🚀 TandemViz v3.7.0 Release Notes. Introducing Our New AI Model: LeadRL
LeadRL; membrane MD; ML-based small-molecule force field; 3D pharmacophore RL; pharmacophore-guided docking; structure prediction upgrades; covalent FEP;
Nov 10 2025
This release brings major new capabilities across TandemViz. We’re excited to introduce LeadRL to TandemFEP. This new AI model uses your FEP calculations to intelligently generate compounds with improved affinity.
New Features & Enhancements
LeadRL
🧠 Physics-Informed Generative Design for Lead Discovery
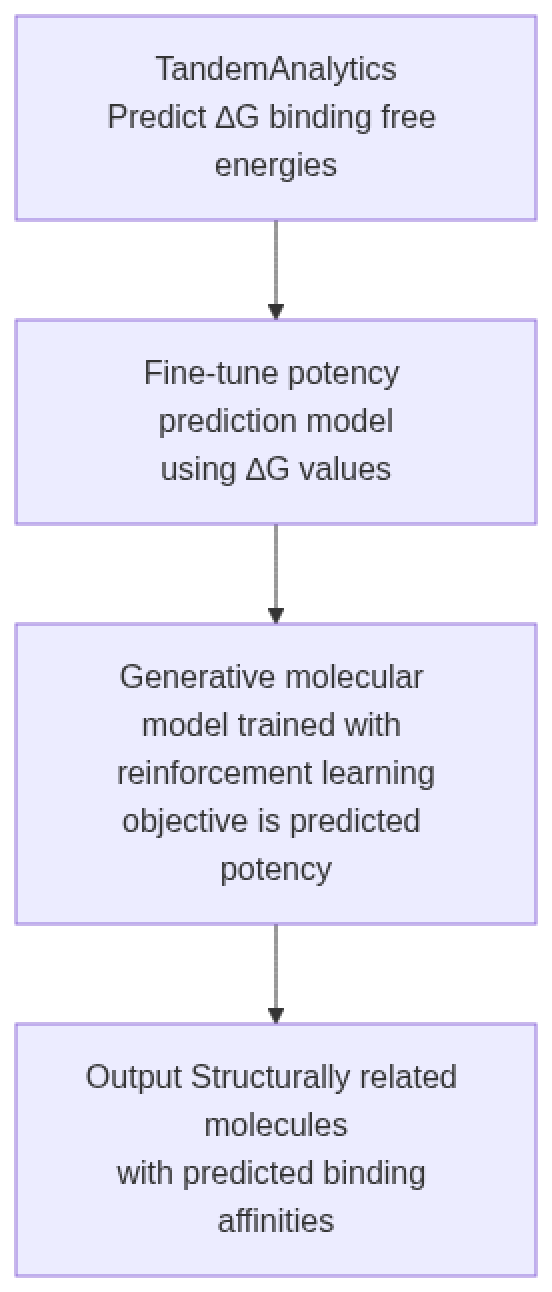
LeadRL uses binding free energies (ΔG) calculated with FEP to fine-tune a 3D structure–based potency model on your specific target and ligand series, capturing both global activity trends and pocket-level SAR. A generative AI model with a prior trained for small molecule lead optimization is then iteratively updated via reinforcement learning using this model fine-tuned with FEP as the reward function. The optimized model can then be used to rapidly generate small molecule analogues with higher affinity leveraging all the data from the FEP calculations.
👉 Try the interactive tutorial
LeadRL: How It Works
LeadRL automatically runs on your TandemFEP results and is included as part of TandemAnalytics
TandemDynamics
This release expands the types of systems you can simulate, adds membrane support across key modes, and makes restraints easier to reuse.
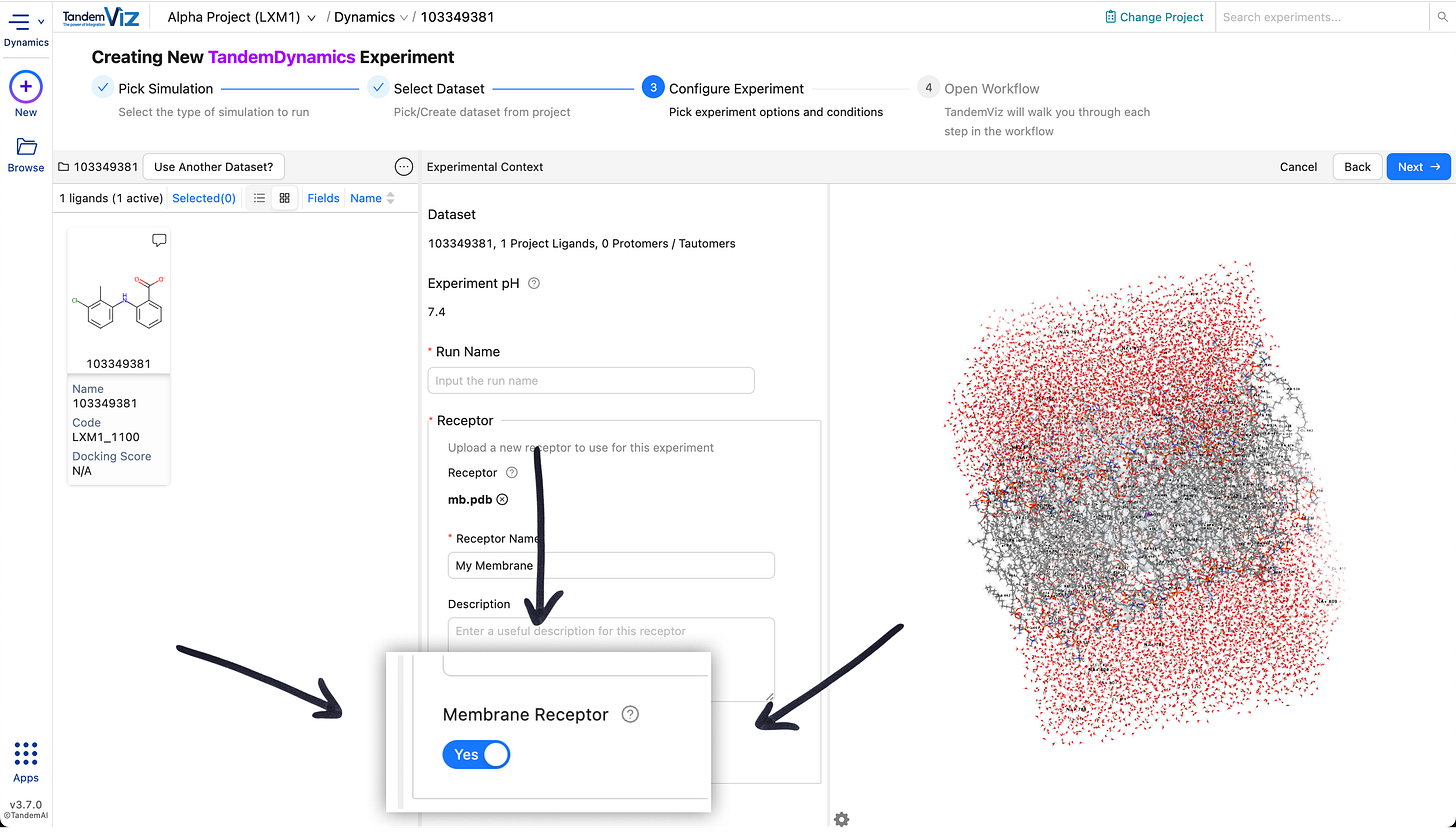
🧬 Membrane Simulations
Membrane systems are now supported for both protein–ligand and receptor-only simulations. When creating a TandemDynamics experiment, simply enable “Membrane Receptor” to enter the membrane workflow.
⬇️ Trajectory Downloads for All Modes
Regardless of the simulation type you run — protein–ligand, receptor-only, ligand-only, or membrane systems — trajectories can now be downloaded directly from TandemViz for local analysis or visualisation in external tools.
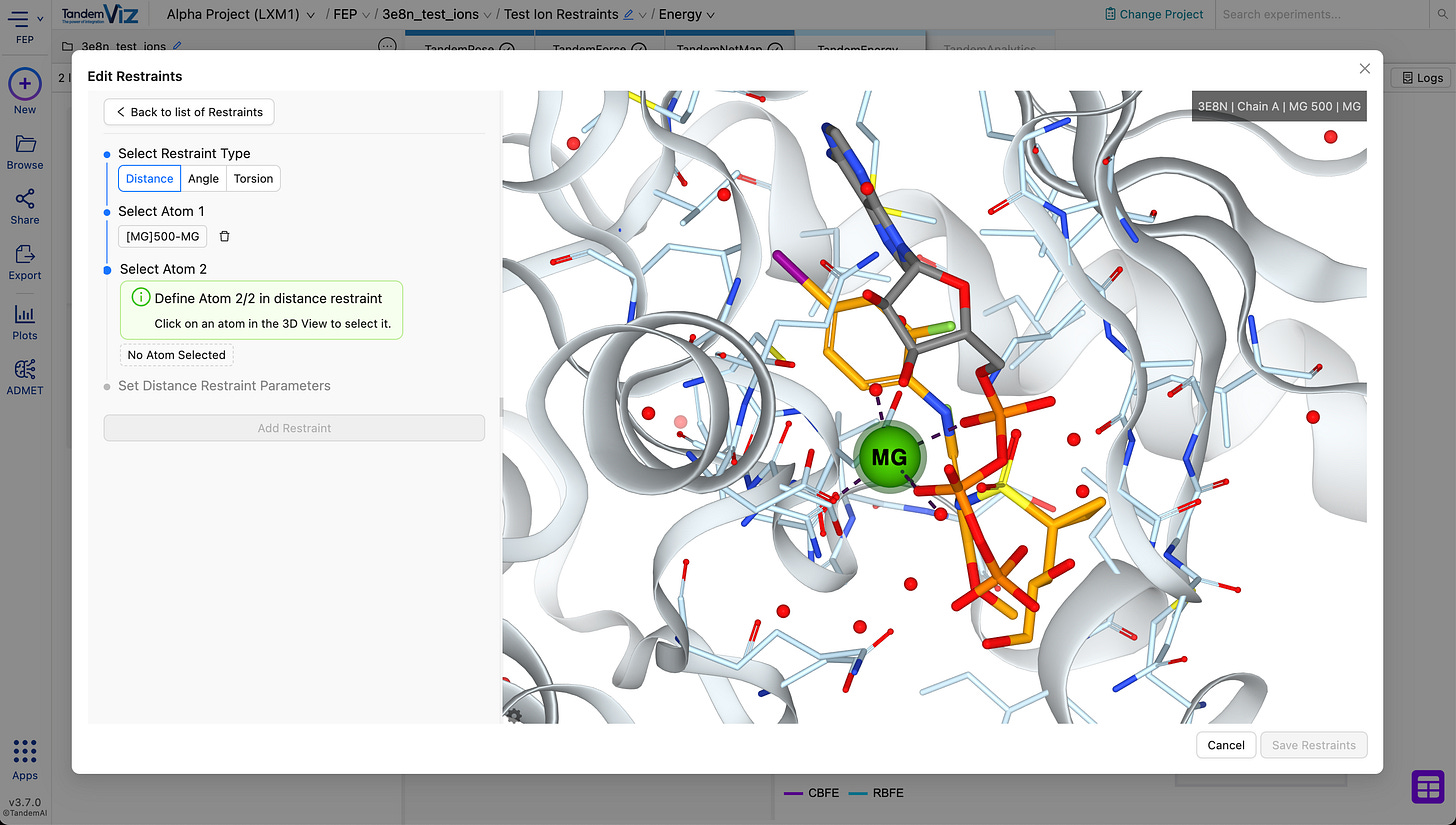
🧲 Ion Cofactor Support in Restraints
Restraints have been extended to support ion cofactors.
📤 Import and Export Restraints
Restraints can now be imported and exported as reusable configuration files. This enables you to quickly transfer restraint definitions between related experiments, standardise restraint usage across a project, or share restraint settings with collaborators.
👉 Try the interactive tutorial
TandemFEP
This release introduces a new ML-based small-molecule force field, plus seamless integration with TandemSandbox.
⚛️ ML-Based Small-Molecule Force Field (TandemForce)
A brand-new method for generating small-molecule force field parameters is now available. Built using a machine-learning model trained on high-level quantum mechanical data, this new force field delivers QM-level precision at dramatically reduced compute cost. It integrates directly into the FEP workflow, with no additional setup required.
🔗 Open TandemAnalytics Frames in TandemSandbox
From TandemAnalytics, you can now open any trajectory frame directly in TandemSandbox for structural inspection. This allows you to perform measurements, add comments, compare poses, and use it in a TandemSandbox session.
🧠 Automatic Mapping Tool in the Atom Mapping Tool
The atom mapping tool just got more intelligent. Use the new automatic mapping tool to rapidly map selection of atoms.
👉 Try the interactive tutorial
TandemGen
This release enhances reinforcement learning capabilities, adds a new 3D pharmacophore objective, and improves sampling, filtering, and brings new ML-ADMET filters.
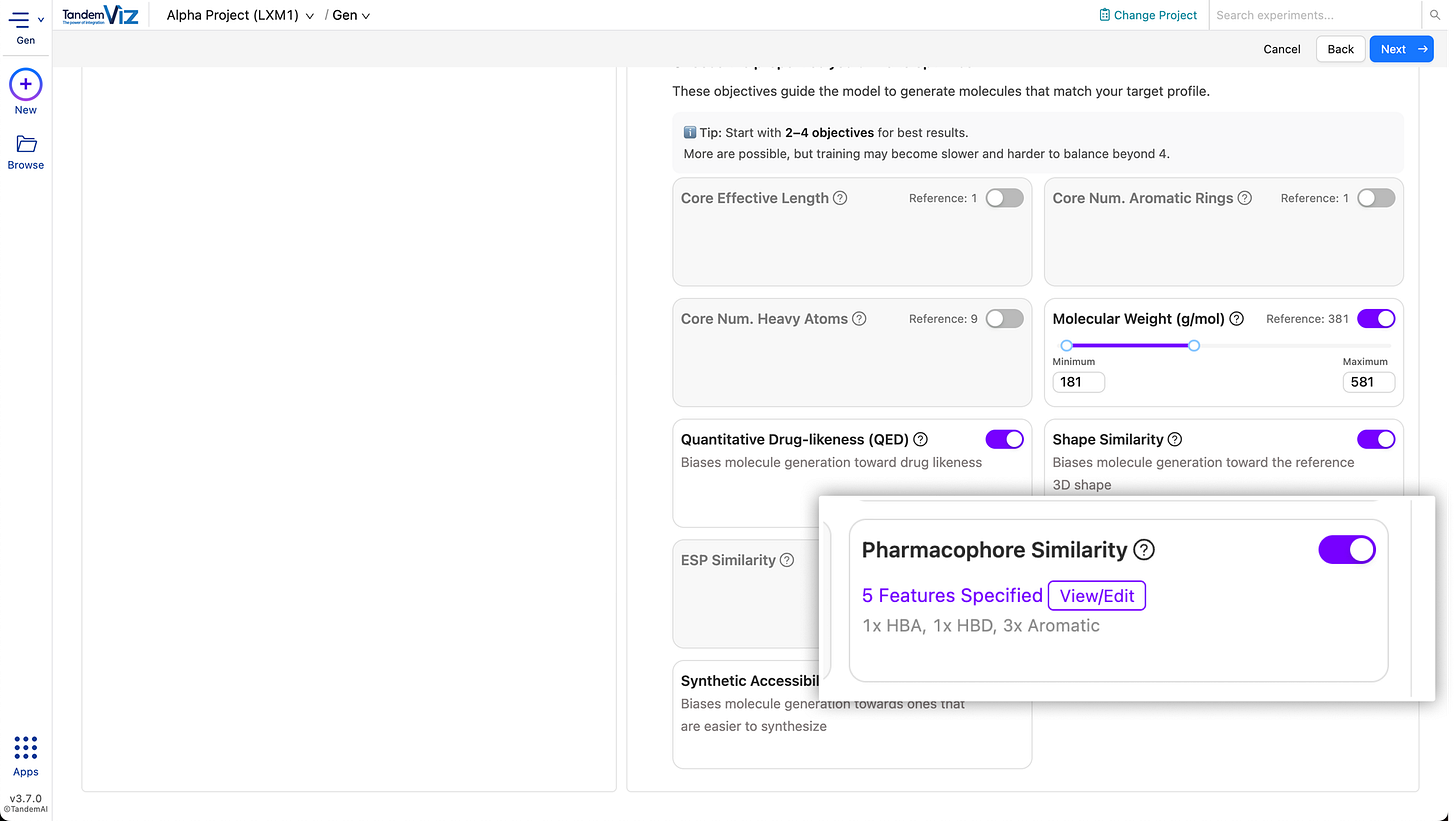
🧲 3D Pharmacophore Objective for RL
Reinforcement learning in TandemGen now supports a 3D pharmacophore objective. You can optimise libraries to satisfy key pharmacophore features, providing an additional structure-aware optimisation strategy beyond shape and ESP.
👉 Try the interactive tutorial
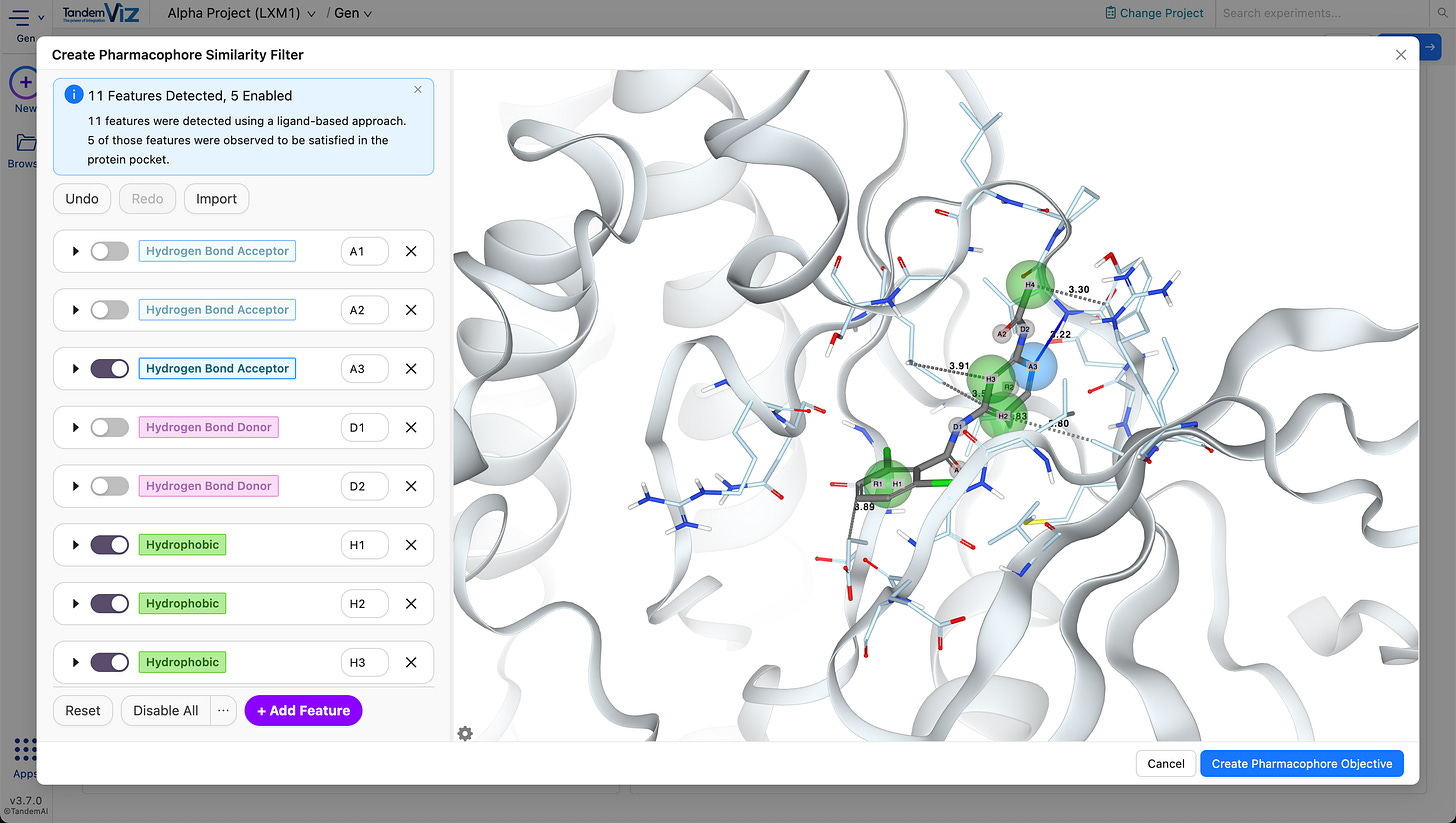
✨ Automatic Pharmacophore Model Generation
3D pharmacophore models can now be generated automatically from the reference ligand and if a receptor is provided, the model is refined to retain only features observed in the binding pocket, making it easy to produce pocket-relevant pharmacophore queries in just a few clicks.
👉 Try the interactive tutorial
➕ “Generate More” Without Retraining
Once the RL model finishes training, you can now sample additional molecules without retraining the model. This enables fast exploration of additional ideas from the same learned distribution, saving time while expanding chemical diversity from successful runs.
⚡ Improved Status Reporting For Task Filters
All asynchronous filters like shape, ESP, and pharmacophore similarity filters now report progress more clearly in the filter.
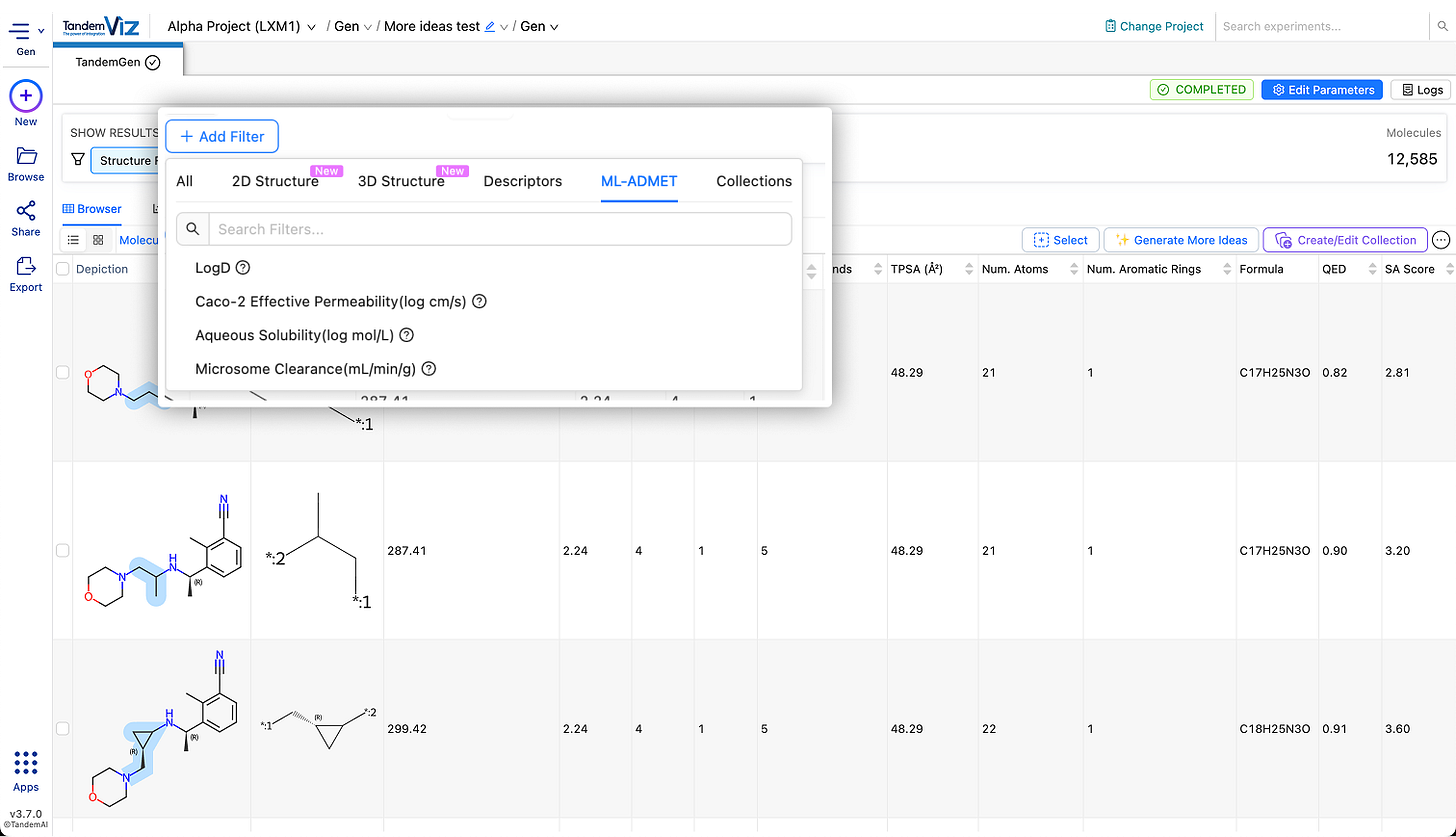
🧪 New ML-ADMET Filters
Four new ML-ADMET properties have been added to the filtering toolkit. They can be applied directly from Add Filter → ML-ADMET to rapidly filter candidates with more favourable ADMET characteristics.
👉 Try the interactive tutorial
TandemDock
This release introduces pharmacophore-guided docking and adds flexibility in engine selection.
🎯 Pharmacophore-Guided Docking
Docking can now be guided using 3D pharmacophore queries. By supplying a pharmacophore model, the docking engine will penalize poses that don’t satisfy the provided features, helping maintain critical interactions during pose generation.
👉 Try the interactive tutorial
🔁 Flexible Engine Selection
Both Vina and rDock are now available for free docking runs.
TandemSandbox
This release makes it easier to launch new experiments directly from your active session and enhances structure prediction.
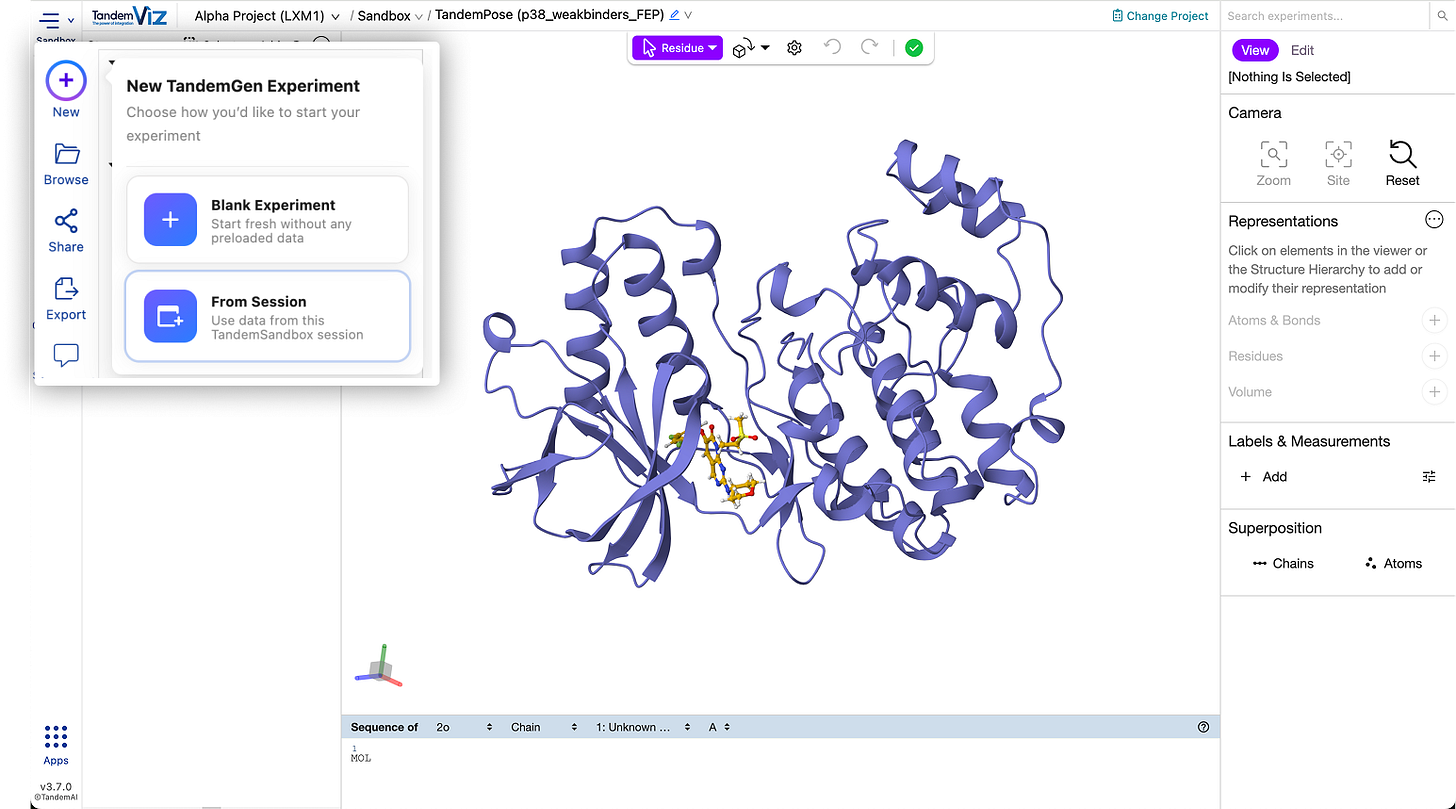
⚡ Start Experiments Directly from Session Assets
You can now start new experiments using the assets currently loaded in your session. For example, when launching a TandemGen run from a co-crystal structure, the reference is automatically pre-selected. This applies across applications and streamlines experiment creation from TandemSandbox.
👉 Try the interactive tutorial
🧬 Structure Prediction Enhancements
Structure prediction now supports the use of templates, improving model accuracy when homologous structures are available. You can also bulk import ligands during setup to run predictions across a series. Prediction details and binding affinity results are available to download in the Sandbox session for easy review.
👉 Try the interactive tutorial
Projects
📊 Plot Any Descriptor or Custom Field
You can now generate plots using any descriptor or custom field imported into TandemViz directly from the project view.