🚀 TandemViz v3.5.0 Release Notes
structure prediction from sequence; 3D pharmacophore filters; open-source RDKit MCP server; random library sampling; .sdf export; rerun FEP edges easily; customizable interaction plots
This release makes TandemGen more powerful than ever, enabling the generation of high-quality, directed, and easily filterable libraries of compounds using 3D pharmacophores.
👉 Access an interactive tutorial showcasing the latest features
We’re also introducing a seamless structure prediction experience, like RosettaFold All-Atom, Boltz2 and other leading models. By combining these tools with TandemSandbox, our collaborative 3D design environment, we believe we’re offering the most intuitive and powerful structure prediction workflow in the industry.
In addition to these major upgrades, we’ve addressed many user requests, resolved key bugs, and made improvements to performance and stability throughout TandemViz.
Major New Features
🧬 Structure Prediction Capabilities – Sequences to Structures in One Step
You can now generate 3D structures directly from protein, RNA, or DNA sequences alongside ligands. The folded complex is opened automatically in TandemSandbox, our collaborative 3D visualization workspace.
Boltz2 is the default
RosettaFold All Atom is enabled out of the box to
👉 Try out an interactive tutorial here
Why Structure Prediction, and Why in TandemSandbox?
Predicting the 3D structure of a protein, RNA, or DNA sequence is often the starting point for modern structure-based drug design. Whether you’re exploring new targets or modelling binding pockets for virtual screening, having access to reliable structural models is critical.
We integrated structure prediction directly into TandemSandbox to make this process seamless, interactive, and collaborative. Users can now go from sequence to folded structure in a few clicks, without leaving the design environment. And because TandemSandbox supports real-time collaboration, teams can explore, annotate, and design around predicted structures together
🎯 Pharmacophore Filters in TandemGen
Upload a reference pose and generate a pharmacophore query in one click. Use this query to dynamically filter down the set of molecules produced by TandemGen.
👉 Try out an interactive tutorial here
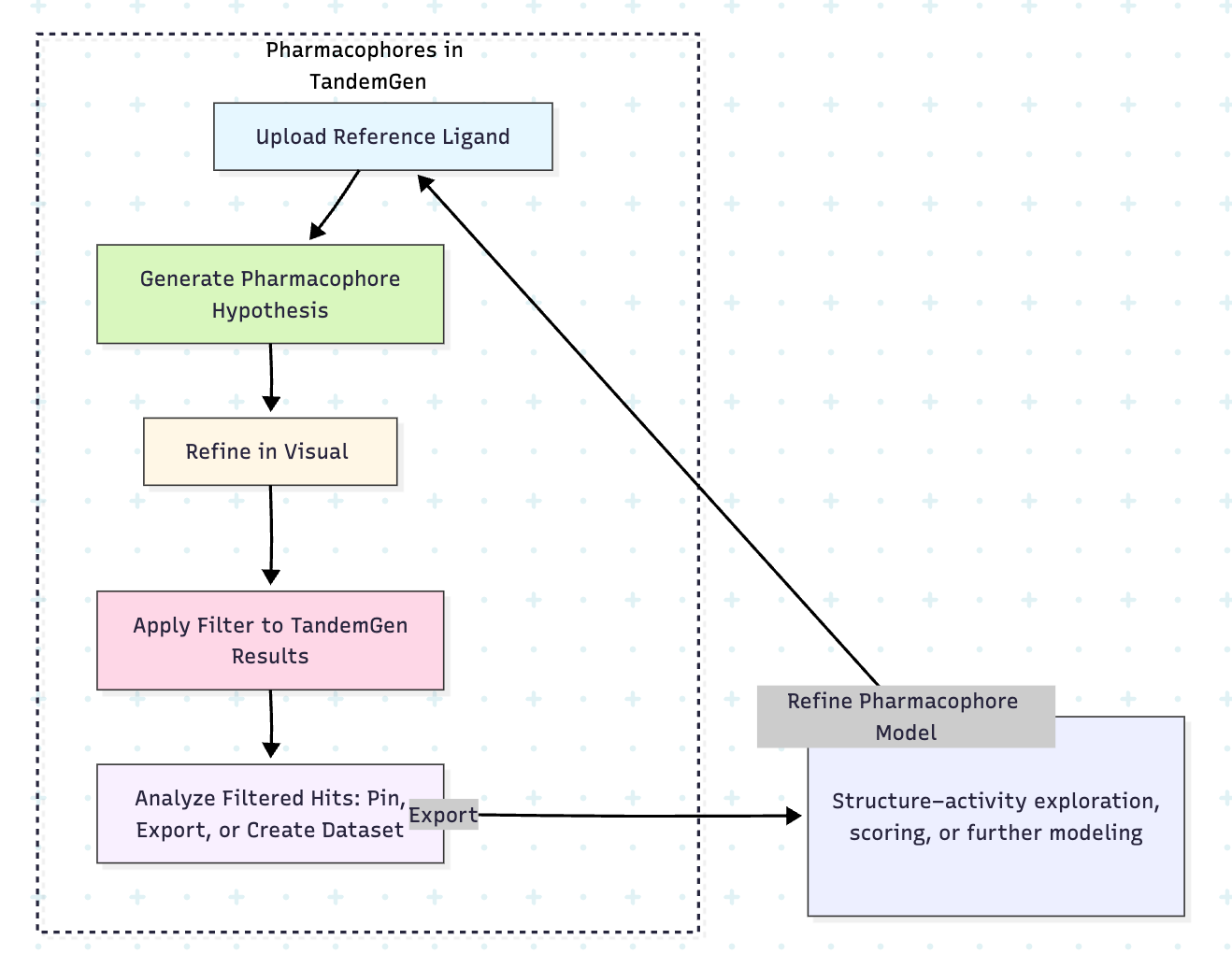
What is a Pharmacophore and Why Does It Matter?
A pharmacophore is a simplified, abstract representation of the key features a molecule needs to interact with a biological target, such as hydrogen bond donors or acceptors, hydrophobic regions, or aromatic features. These features are defined based on the 3D arrangement observed in known active compounds or reference structures.
In the context of TandemGen, pharmacophores are especially powerful: they allow you to filter large, generated libraries to identify molecules that match the spatial and chemical interaction patterns most likely to drive activity. Instead of manually inspecting each molecule, you can now automatically prioritize candidates that align with a desired interaction profile.
✂️ Three-Cut Core Definition
You can now define cores using up to three cuts, giving you greater control over which R-groups are preserved. Previously limited to two cuts, this added flexibility enables more precise scaffold design.
Currently available for the context-aware technique, with support for generative AI-based core hopping coming soon.
🧪 Open-Source RDKit MCP Server
We’ve released the first version of our open-source RDKit MCP server . Designed to allow AI agents to run cheminformatics tasks using RDKit.
By open-sourcing our RDKit MCP server, we’re making it easier for the community to build intelligent, chemistry-aware systems that can query, transform, and analyze molecules at scale. If you’re building LLM agents this server offers a lightweight, flexible way to access RDKit remotely. We invite contributions to help it grow!
New Features & Enhancements
TandemGen
🧪 New Reactions: Urea Formation & Sonogashira Coupling
Two new reactions have been added to the default TandemGen reaction library:
Urea Formation
Sonogashira Coupling
🎯 Reference Values in Reinforcement Learning Objectives
When configuring generative objectives, users now see reference values next to each field. This helps calibrate design goals based on the provided reference
🔀 Random Sampling from Libraries
Quickly sample a random, diverse subset from any enumerated library for faster visualization or selection.
📂 Improved Metadata in Run Browser
We’ve improved the metadata shown for each TandemGen run, making it easier to recall experiment context and navigate past results. The size of the library is now more detailed (post-TandemFilter and total) and includes information on collections and filters.
🔍 Search Existing Ligands to Create Datasets
You can now search previously generated ligands when creating a dataset, in addition to uploading or sketching new structures.
📤 Export to 3D SDF
Easily export 3D molecules from a TandemGen session to a 3D SDF file for downstream use.
👉 Try out an interactive tutorial here
🧱 Unified Dataset Creation Flow
We’ve harmonized the dataset creation experience across TandemViz, making it easier to work consistently between apps.
TandemFEP
🔄 Rerun Edges from Results Table
You can now select and rerun edges directly from the results table, speeding up iteration and refinement of your FEP network.
📊 Export Interaction Data as CSV
Interaction data can now be exported to CSV format, enabling custom offline analysis or integration with your own scripts.
TandemAnalytics
📊 Customisable Interaction Plots
Users can now reorder residues and ligands in interaction heatmaps to tailor visuals for storytelling, reporting, or publication.
TandemADMET
💬 Improved Help Descriptions
We’ve rewritten many help tooltips and field descriptions in TandemADMET to make interpreting results more intuitive and user-friendly.
🛠️ Bug Fixes
Consistent Units and Data Across Tables
Units and data formatting are now consistent throughout all result and summary tables.
Buchwald–Hartwig Reaction Fix (TandemGen)
This reaction now works correctly and outputs the expected results.
Corrected Fragment 3D Geometries (TandemForce)
Fragment geometries are now computed and exported with accurate 3D coordinates.
Thank you for using TandemViz and helping us build a smarter, more collaborative drug discovery platform. As always, we welcome your feedback and look forward to hearing how these new features are helping your team.
— The TandemViz Product Team
















